- Home
- News
- Genes associated with stress tolerance, oil content, seed quality and ecotype improvement were identified by GWAS in rapeseed
Genes associated with stress tolerance, oil content, seed quality and ecotype improvement were identified by GWAS in rapeseed
GWAS
Title: Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement
Journal: Nature Communications
NGS | WGS | Resequencing | GWAS | Transcriptome | RNAseq | Brassica napus | Evolution | Domestication
In this study, Biomarker Technologies provided Services on NGS sequencing, as well as technical support on bioinformatics analysis on sequencing data.
Background
Brassica napus (rapeseed) is an important oilseed crop and an excellent model for investigating processes of polyploid speciation, evolution and selection. However, whether wild species or domesticated donors were parental progenitors and genes that contributed to domestication and improvement in rapeseed are still unknown.
Materials and methods
Materials: 588 B. napus accessions were involved in this study, including 466 from Asia, 102 from Europe, 13 from North America, and 7 from Australia. Based on growth habit records, these materials were divided into three ecotypes; spring (86 accessions), winter (74 accessions), and semi-winter (428 accessions).
Sequencing: Averaged approx. 5× (ranging from 3.37× to 7.71×)
Sequencing platform: Illumina Hiseq 4000
Data production: 4.03 Tb clean data
SNP calling: BWA + GATK. 5,294,158 SNPs and 1,307,151 InDels were obtained.
Results
Origin of B. napus
Fig. 2 Population structure of 588 B. napus accessions and 199 of B. rapa accessions.
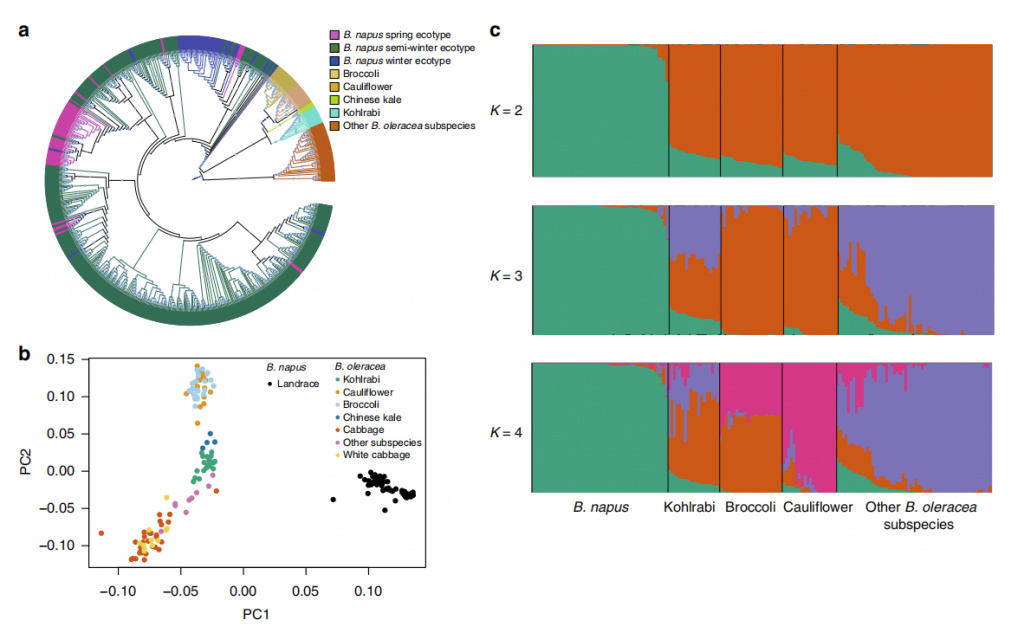
Fig. 3 Population structure of 588 B. napus accessions and 119 of B. oleracea accessions
Selection signals and Genome-wide association studies.
During the first stage of improvement (FSI), more genetic diversity was lost in the B. napus C subgenome than in the A subgenome. Less genetic differentiation occurred during the FSI than during the second stage of improvement (SSI). Genes in SSI-selection signal regions were enriched in stress tolerance, development and metabolic pathways. 60 loci significantly associated with 10 target traits, including 5 related to seed yield, 3 to silique length, 4 to oil content, and 48 to seed quality were identified.
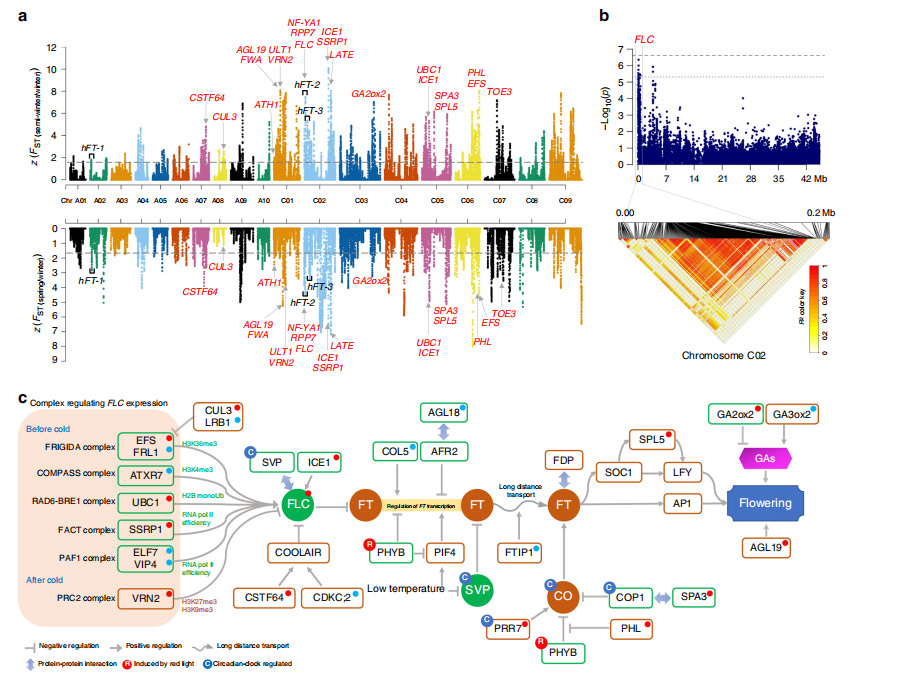
Fig. 4 Genome-wide scanning and annotations of selected regions during the SSI of B. napus
Transcriptome analysis
RNAseq data of 11 tissues from a high-oil-content and double-low cultivar and a low-oil-content and double-high cultivar identified genes related to glucosinolate biosynthetic process were significantly over-represented.
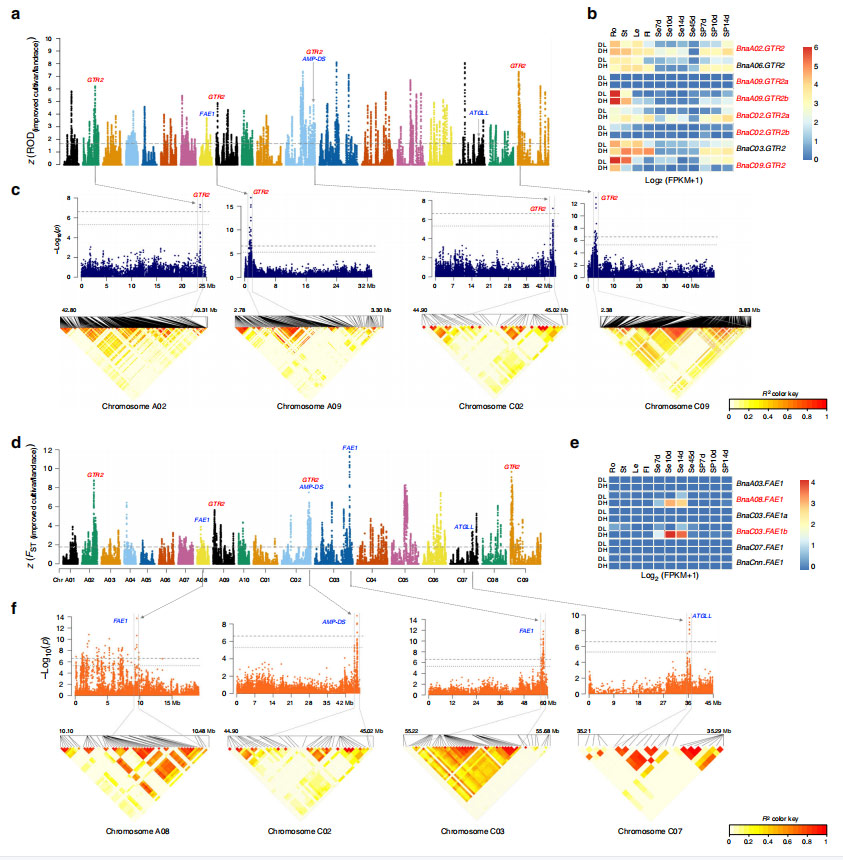
Fig. 5 Overview of flowering-time regulation under the selection of the ecotype improvement of B. napus
Discussion
This study provided a valuable resource for understanding the origin and improvement history of B. napus and will facilitate the dissection of the genetic bases of important agronomic complex traits. The significant SNPs associated with favorable variants, selection signals and candidate genes will largely contribute in future, especially in improving the yield, seed quality, oil content, and adaptability of this recent allopolyploid crop and its relatives.
Reference
Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement[J]. Nature Communications, 2019, 10(1).
News and Highlights aims at sharing the latest successful cases with Biomarker Technologies, capturing novel scientific achievements as well as prominent techniques applied during the study.