- Home
- News
- Structure variants in Chinese population and their impact on phenotypes, diseases and population adaptation
Structure variants in Chinese population and their impact on phenotypes, diseases and population adaptation
WHOLE GENOME RESEQUENCING

Structure variants in Chinese population and their impact on phenotypes, diseases and population adaptation
Nanopore | PacBio | Whole genome re-sequencing | Structural variation callling
In this study, Nanopore PromethION sequencing was provided by Biomarker Technologies.
Highlights
In this study, an overall landscape of structural variations(SVs) in human genome was revealed with help of long-read sequencing on Nanopore PromethION platfrom, which deepens the understanding of SVs in phenotypes, diseases and evolution.
Experimental Design
Samples: Peripheral blood leukocytes of 405 unrelated Chinese individuals(206 males and 199 females) with 68 phenotypic and clinical measurements. Among all individuals, ancestral regions of 124 individuals were provinces in North, those of 198 individuals were South, 53 were SouthWest and 30 were not known.
Sequencing strategy: Whole genome long-read sequencing(LRS) with Nanopore 1D reads and PacBio HiFi reads.
Sequencing platform: Nanopore PromethION; PacBio Sequel II
Structure Variation Calling
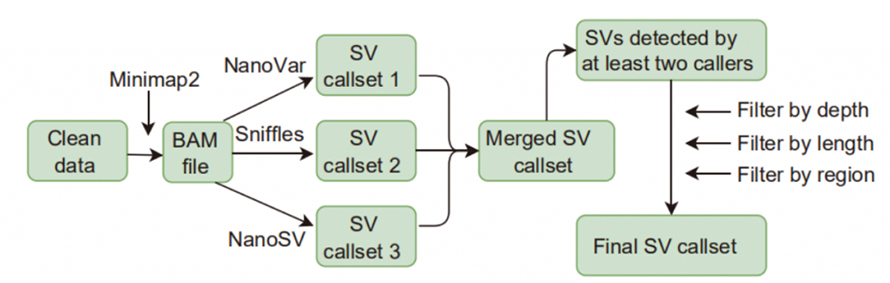
Figure 1. Workflow of SV calling and filtering
Main Achievements
Structure variation discovery and validation
Validation by PacBio: SVs identified in one sample(HG002, child) were validated by a PacBio HiFi dataset. The overall false discovery rate(FDR) was 3.2%, illustrating a relatively reliable SV identification by Nanopore reads.
Non-redundant SVs and genomic features
Genomic features: Number of SVs was found significantly correlated with chromosome length. Distribution of genes, repeats, DELs(green), INS(blue), DUP(yellow) and INV(orange) were displayed on a Circos diagram, where a general increase in SV were observed at the end of chromosome arms. (Figure 3d and 3e)
Length of SVs: Lengths of INSs and DELs were found to be significantly shorter than those of DUPs and INVs, which agreed with those identified by PacBio HiFi dataset. Length of all identified SVs added upto 395.6 Mb, which occupied 13.2% of entire human genome. SVs affected 23.0 Mb(approx. 0.8%) of genome per individual in average. (Figure 3f and 3g)
Functional, phenotypical and clinical impacts of SVs
A common DEL of 2.4 kb was observed in 35 homozygous and 67 heterozygous carriers, which covers the complete region of the 3rd exon of Growth Homone Receptor(GHR). The homozygous carriers were found significantly shorter than heterzygous ones(p=0.033). (Figure 4d)
Furthermore, these SVs were processed for population evolutionary studies between two regional groups: North and South China. Significantly differential SVs were found distributed on Chr 1, 2, 3, 6,10,12,14 and 19, within which, top ones were associated with immunity regions, such as IGH, MHC, etc. It is reasonable to spectulate that the differentiation in these SVs may due to genetic drift and long-term expose to diverse envronments for sub-populations in China.
Reference
Wu, Zhikun, et al. “Structural variants in Chinese population and their impact on phenotypes, diseases and population adaptation.” bioRxiv (2021).
News and Highlights aims at sharing the latest successful cases with Biomarker Technologies, capturing novel scientific achievements as well as prominent techniques applied during the study.