- Home
- News
- A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes
A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes
GENOME EVOLUTION
nature genetics
A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes
PacBio | Illumina | Bionano Optical Map | Hi-C Genome Assembly | Genetic Map | Selective Sweeps | RNA-Seq | ISO-seq | SLAF-seq
Biomarker Technologies provided technical support on Pacbio sequencing, Hi-C sequencing and data analysis in this study.
Highlights
1.The first chromosomal-level high quality Rye genome was obtained, which has a single chromosome size larger than 1 Gb.
2.Compared to Tu, Aet and Hv genome, a unique recent LTR-RT events was observed in Rye genome, which was responsible for the extension of the rye genome size.
3.The divergence between rye and diploid wheats took place after the separation of barley from wheat, with the divergence times for the two events being approximately 9.6 and 15 MYA .
FT genes phosphorylation may control the early heading trait in rye.
4.Selective sweep analysis indicate possible involvement of ScID1 in the regulation of heading date and its probable selection by domestication in rye
Background
Background
Rye is a valuable food and forage crop, an important genetic resource for wheat and triticale improvement, and an indispensable material for efficient comparative genomics studies in grasses. Weining rye, an early flowering variety cultivated in China, is outstanding because of its broad-spectrum resistance to both powdery mildew and stripe rust. To understand the genetic and molecular basis of rye elite traits and to promote the genomic and breeding studies in rye and related crops, we here sequenced and analyzed the genome of Weining rye.
Achievements
Rye Genome
The Rye genome was constructed by combing PacBio SMRT reads, short-read Illumina sequencing, as well as those from chromatin conformation capture (Hi-C), genetic mapping, and BioNano analysis. The assembled contigs (7.74 Gb) accounted for 98.47% of the estimated genome size (7.86 Gb), with 93.67% of the contigs (7.25 Gb) assigned to seven chromosomes. Repetitive elements constituted 90.31% of the assembled genome.
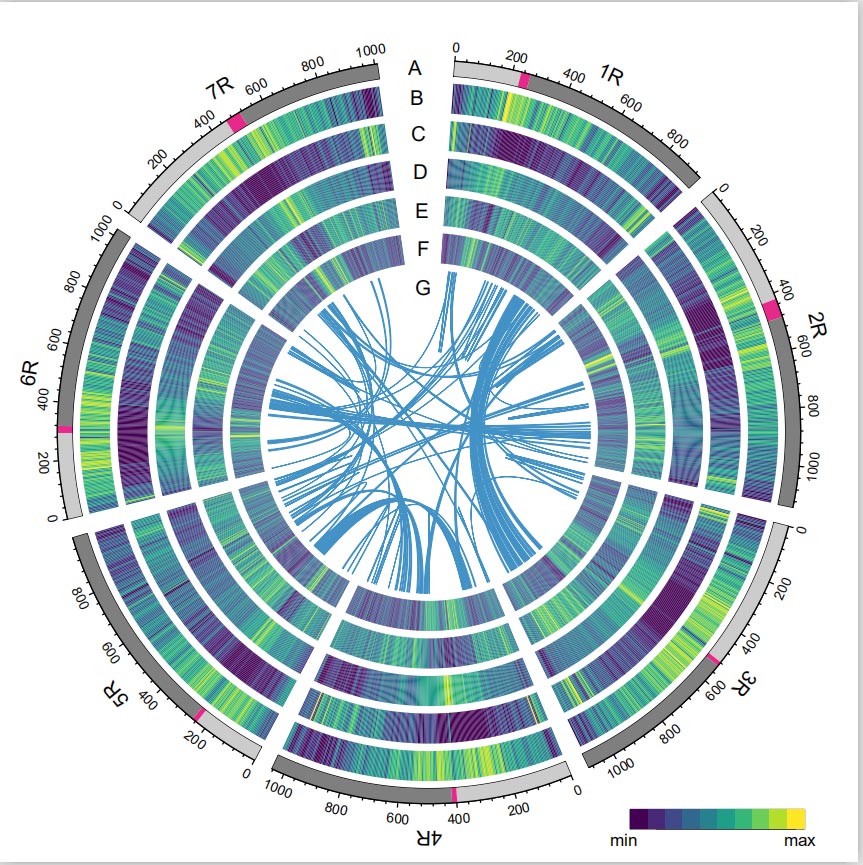
Rye Genome
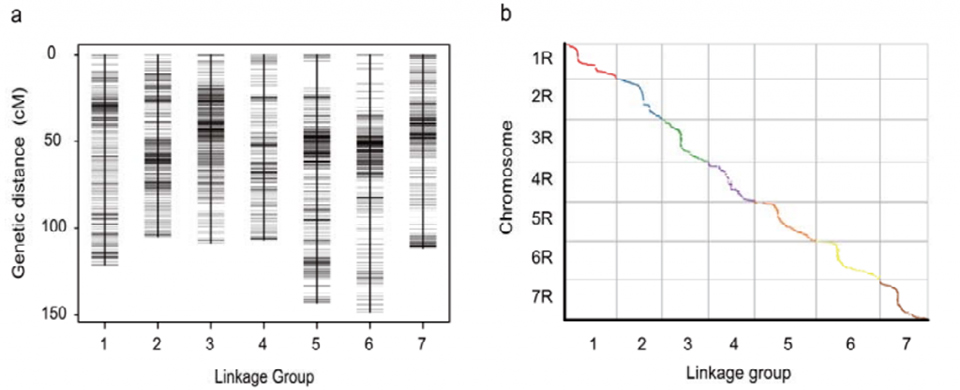
Genetic linkage map (WJ) developed using 295 F2 plants derived from the crossing of two rye landraces (Weining × Jingzhou)
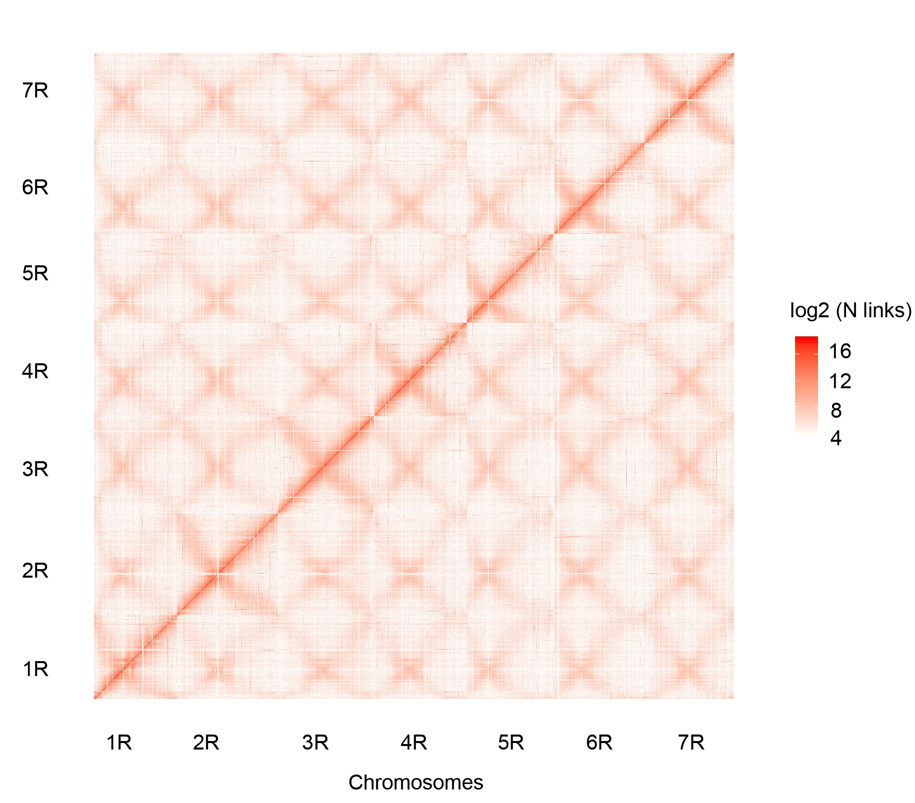
Hi-C contact map of the seven assembled Weining rye chromosomes (1R – 7R)
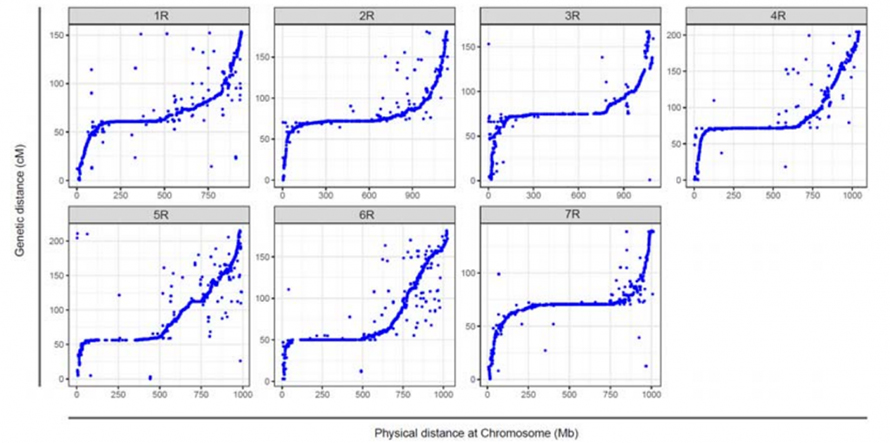
Alignment between the seven assembled chromosomes of Weining rye and the seven rye linkage groups developed using Lo7 x Lo255 RIL population
The LTR Assembly Index (LAI) value of the Rye genome was found to be 18.42 and 1,393 (96.74%) of the 1,440 highly conserved BUSCO genes were identified out .These results suggest that the Weining rye genome sequence is of high quality in both intergenic and genic regions. A total of 86,991 protein-coding genes, including 45,596 high-confidence(HC) genes and 41,395 low-confidence genes (LC) genes were predicted.
2. Analysis of TEs
Analysis of TEs. A total of 6.99 Gb, representing 90.31% of Weining assembly, was annotated as TEs, which included 2,671,941 elements belonging to 537 families. This TE content was clearly higher than that previously reported for Ta (84.70%), Tu (81.42%), Aet (84.40%), WEW (82.20%), or Hv (80.80%). The long terminal repeat retrotransposons (LTR-RTs), including Gypsy, Copia and unclassified RT elements, were the dominant TEs, and 1ccupied 84.49% of annotated TE content and 76.29 % of assembled Weining genome; CACTA DNA transposons were the second most abundant TEs, constituting 11.68% of annotated TE content and 10.55 % of assembled Weining genome.
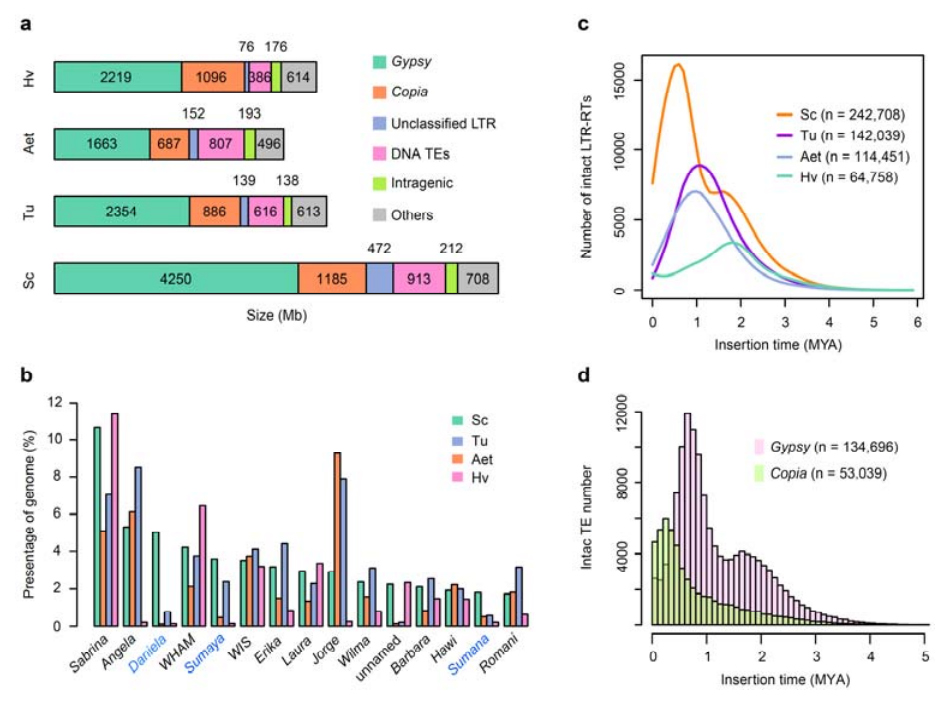
Analysis of transposon elements of rye
Weining rye had a comparatively high proportion of recent insertions of LTR-RTs with the peak of amplification appeared around 0.5 million years ago (MYA), which was the most recent among the four species; the other peak, occurred approximately 1.7 MYA, was older and also seen in barley. At superfamily level, very recent bursts of Copia elements in Weining rye at 0.3 MYA was found, while the amplifications of Gypsy RTs dominantly shaped the bimodal distribution pattern of LTR-RT burst dynamics.
3. Investigation of rye genome evolution and chromosome syntenies
The divergence between rye and diploid wheats took place after the separation of barley from wheat, with the divergence times for the two events being approximately 9.6 and 15 MYA, respectively. 1R, 2R, 3R were entirely collinear with groups 1, 2 and 3 chromosomes of wheat, respectively. 4R, 5R, 6R, 7R were found exists large-scale fusions and segments.
4. Analysis of gene duplications and their impact on starch biosynthesis genes
Notably, the numbers of tandemly duplicated genes (TDGs) and proximally duplicated genes (PDGs) of Weining rye were both higher than those found for Tu, Aet, Hv, Bd and Os. Transposed duplicated genes(TrDGs) were also more numerous than those specifically found for Tu and Aet. Rye genome expansion is accompanied by higher numbers of gene duplications. The increased TE bursts in rye may have led to an elevated number of TrDGs.
5. Dissection of rye seed storage protein (SSP) gene loci
Four chromosomal loci (Sec-1 to Sec-4) specifying rye SSPs have been identified on 1R or 2R. α-gliadin genes were evolved only recently in wheat and closely related species after the divergence of wheat from rye.
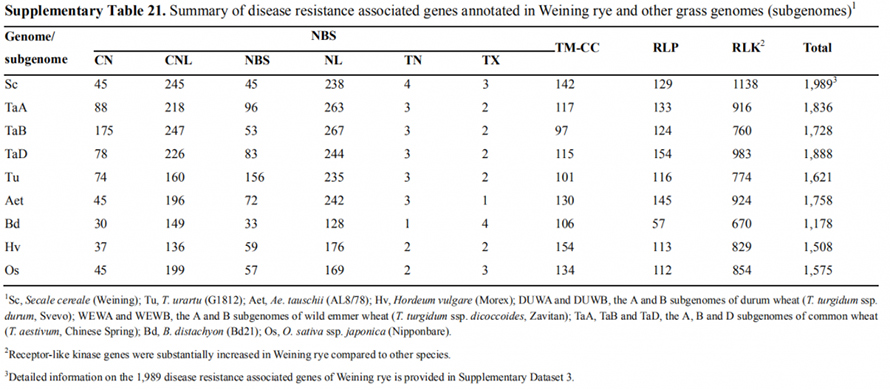
6. Examination of transcription factor (TF) and disease resistance genes
Weining rye had more numerous disease resistance associated (DRA) genes (1,989, Supplementary Data 3) than Tu (1,621), Aet (1,758), Hv (1,508), Bd (1,178), Os (1,575), and the A (1,836), B (1,728) and D (1,888) subgenomes of common wheat.
7. Investigation of gene expression features associated with early heading trait
Two FT genes with relatively high expression under long day conditions, ScFT1 and ScFT2 , were annotated in Weining genome assembly. Two amino acid residues of ScFT2 (S76 and T132) phosphorylation were found relationship with lowering time control
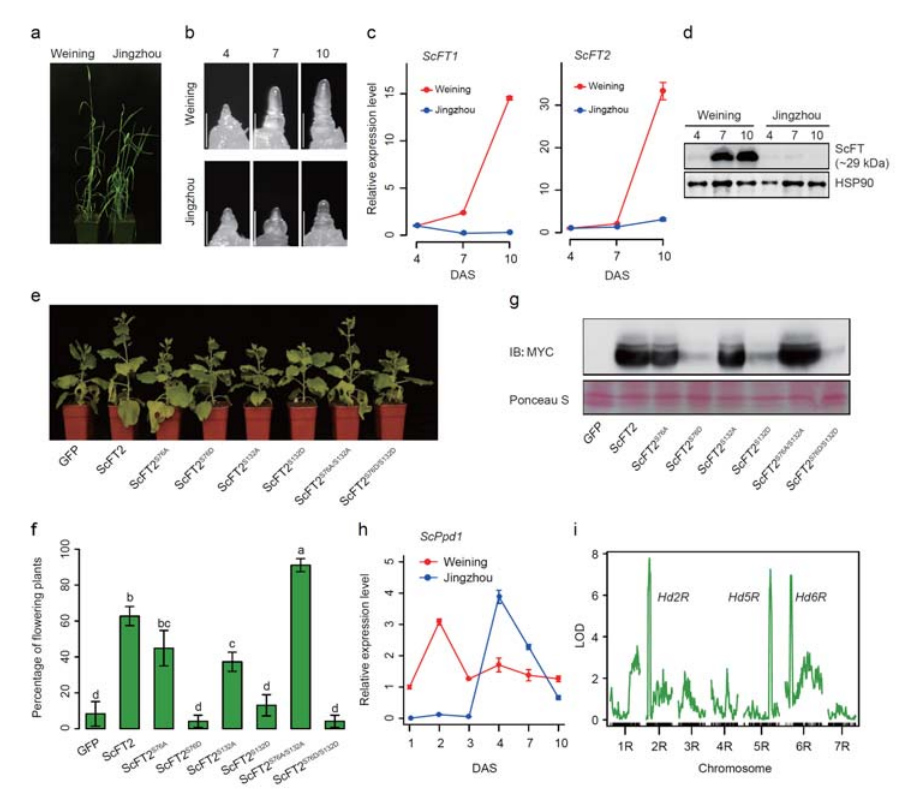
Developmental and gene expression features associated with the early heading trait of Weining rye
8. Mining of chromosomal regions and loci potentially involved in rye domestication
A total of 123,647 SNPs were used to conduct selevtive sweep analysis between cultivated rye and S. vavilovii. 11 selective sweep signals identified by reduction index (DRI),fixation index (FST) and XP-CLR method. ScID1 was found possible involvement in the regulation of heading date.
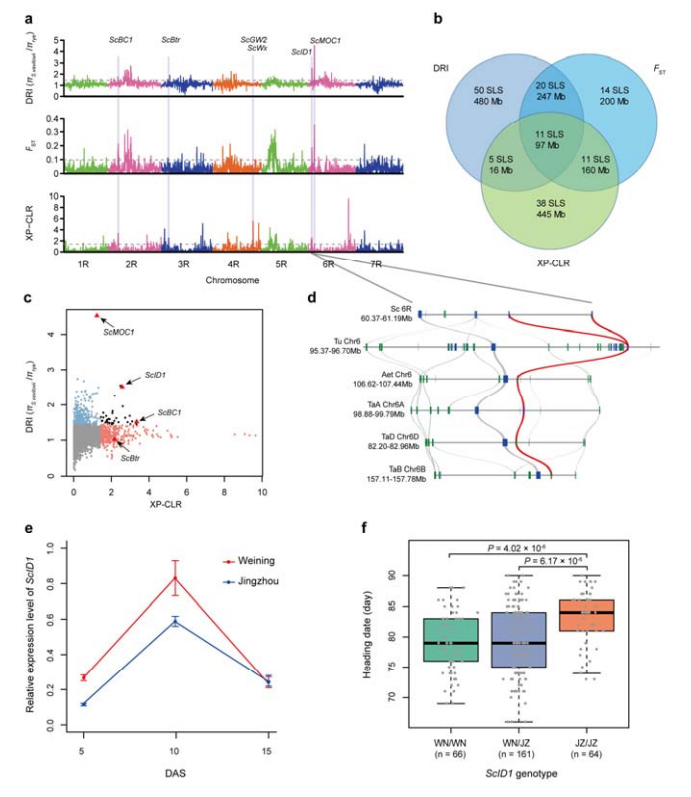
Identification and analysis of chromosomal regions and loci potentially related to rye domestication
Reference
Li G.W. et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nature Genetics (2021)
News and Highlights aims at sharing the latest successful cases with Biomarker Technologies, capturing novel scientific achievements as well as prominent techniques applied during the study.